Another genealogy for SARS-CoV2 genomes
Over at Peaceful Science, Josh Swamidass has called attention to two studies that inferred genealogical trees for the SARS-CoV2 virus using molecular phylogeny methods. Here is an image of one of them, inferred 3 weeks ago by Andrew Rambaut, a major researcher in molecular evolution of viruses:
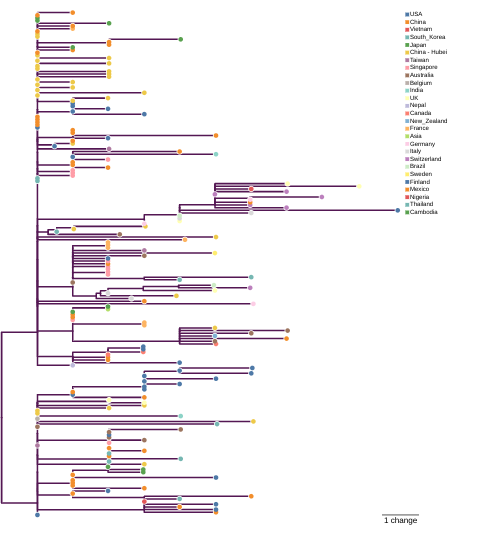
(This is a screenshot – the Rambaut article with its interactive figures can be found here). We’ve seen here in my earlier post a tree of genomes produced by Trevor Bedford of the Fred Hutchinson Cancer Research Center here in Seattle. I haven’t compared them yet, so maybe you should do that. I know both these guys and they both really know what they are doing. I would expect both trees to show quite compatible structure (keep in mind that they will not contain exactly the same genomes as they were produced at different times, so we have to look at the branches connecting the genomes that they have in common).
Rambaut’s twitter feed and article also include resources on phylogeography – the geographic distribution of the genealogical tree.
They both try to estimate when the epidemic started. I think that Andrew’s tree estimates the time of the first fork in the tree,
while Trevor tries to estimate when the first genome entered humans. On a quick read these come to quite consistent results, the former
a little later than the latter. If
you want to explore their findings further look at their Twitter feeds, which can be found here:
As a bonus I include a link to my colleague Carl Bergstrom’s Twitter feed – he was, I think, the original popularizer of “Flatten The Curve”, and his figure on that was retweeted by President Obama. He is not only an expert in mathematical modeling of information transmission in biology and on the internet, but has been working seemingly tirelessly to debunk wrong arguments online that seek to minimize our response to the pandemic.
