A visit from the architects of the fly hourglass paper
So, shortly after the [first post on the hourglass model](http://pandasthumb.org/archives/2010/12/its-just-a-stag.html) went up here on the Panda's Thumb, the senior author of one of the two featured papers (the article using fruit flies, titled "Gene expression divergence recapitulates the developmental hourglass model") contacted me, clearly enthused about our interest in the story. He's [Pavel Tomancak](http://www.mpi-cbg.de/en/research/research-groups/pavel-tomancak/pi.html), and together with the co-first author on the study, Alex Kalinka, he offered some useful feedback as well as some cool images. Here are some of their further thoughts, posted with their permission and edited slightly by me. Let's think of them as honored guest bloggers.
---------------
I am indeed excited about the insightful blogging that has followed the publication of our work. It is great to see that people like you are genuinely trying to come to grips with the sometimes complex analysis we have put forward (a 55 page monograph accompanies the paper) and share their understanding with scientifically-inclined readers on the blogosphere.
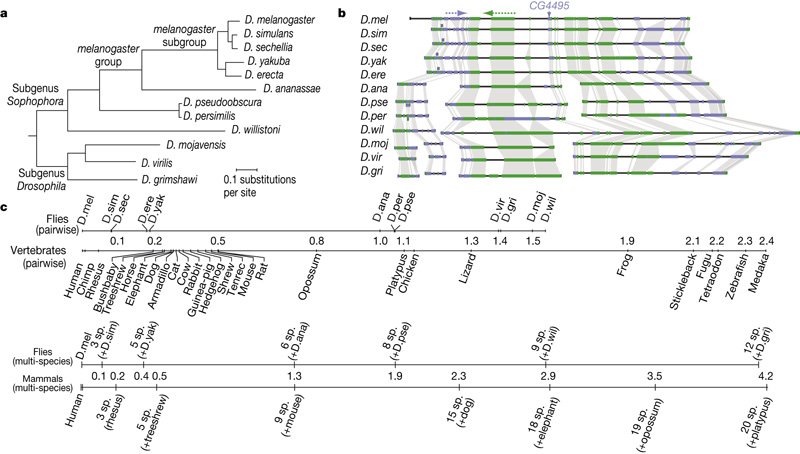
The major challenge we had to overcome was to convince people that the evolutionary divergence of the _Drosophila_ species we used is sufficient to make arguments about phylum-level morphological patterns. Many people thought that we could not possibly say something about the phylotypic stage by looking at closely related species. We made a number of arguments like: the species facilitate unambiguous comparisons, gene orthology is clear, timing of development is not so different (although we compensated for that) and so on. What really drives it home is the comparison of the _Drosophila_ phylogeny to the vertebrate one. _D. virilis_ is roughly as distant from _D. melanogaster_ as lizards are from humans in terms of the molecular clock (see figure below, from [Stark et al. 2007 Fig. 1](http://www.nature.com/nature/journal/v450/n7167/fig_tab/nature06340_F1.html)). I take it that flies are more similar to each other morphologically than say human or mouse or elephant, but they also are rather different, living in very different environments that influence their embryonic strategies (Markow & O'Grady 2008).
However, in addition to these arguments, we believe that approaching evo-devo questions using a mixture of closely related species, such as _D. persimilis_ and _D. pseudoobscura_, together with more distantly related species such as _D. virilis_, allows us to see both large accumulated differences between species that have accrued over tens of millions of years, together with the first steps that occur as two species diverge from one another. One of the advantages of such an approach is that it will enable us to infer whether most differences are the result of a large number of small changes, or whether they are the result of a small number of large changes. Moreover, this approach is essential if we are to integrate population genetics theory with comparative embryology, a synthesis that will greatly advance our understanding of how development evolves (Simpson 2002).
The dataset was actually massive even by today's standards; we had replicates which allowed us to do all the statistics. All together 200 array experiments on self-designed arrays.
Our ability to recapitulate the known phylogeny from the divergence data is in my opinion remarkable. As far as we know, the reconstruction of a phylogeny using expression data has not been attempted before. Karolina Varga, co-first author on the paper, with her impeccable sense of humor, wanted to coin the term 'expressogram' as a variation of 'phylogram', which follows the fine tradition of ‘eisengrams’ and such. Despite exhibiting the same topology as the molecular clock phylogram there are differences (See supplementary Figure 3 of our paper, included below) and these pointed us to the evolutionary modeling that suggests that the hourglass waist is maintained by stabilizing selection.
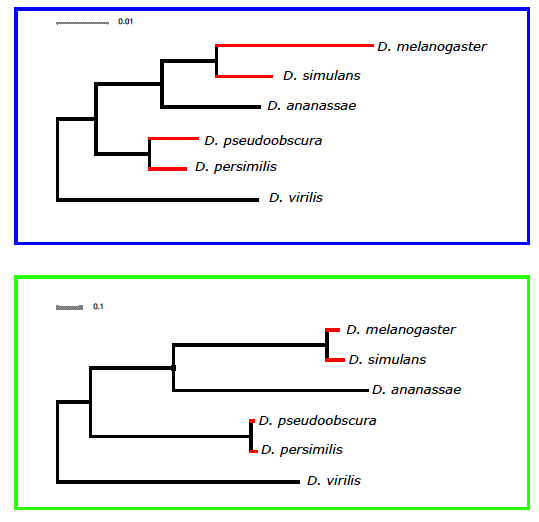
Supplementary Figure 3. A comparison of the phylogenetic relationships between the six Drosophila species used in our study when drawn up using our gene expression data (based on normalised values from a linear model (see Methods)) - boxed in blue - and based on median dS values for ~ 10,000 orthologous genes - boxed in green. Red branches indicate terminal branches between sister species that are longer in the gene expression data.
The genes that conform to the hourglass pattern are enriched for genes involved in building up the body plan and could be in general referred to as 'developmental toolkit genes'. It will be interesting to compare these genes to the ones that drive the hourglass pattern in [the other study](http://www.nature.com/nature/journal/v468/n7325/full/nature09632.html) by Domazet-Loso and Tautz. While there may be commonalities in the studies, it may also be the case that the evolution of new genes (through a process of duplication and divergence, for example) may be under a different set of constraints than the evolution of gene expression. Exploring such differences promises to be an exciting avenue for future research.
Finally, I think that both papers really open up new horizons for studying the developmental hourglass experimentally using the awesome power of _Drosophila_ and fish genetics. That's what we worked towards from the beginning. We are developing fancy transgenic, imaging and image-analysis techniques to be able to compare gene expression in the different species quantitatively on the spatio-temporal level. It's well-funded (among others by [HFSP](http://www.hfsp.org/) and [ERC](http://erc.europa.eu/)) and these papers will be a huge boost to not only us but the entire field, which until recently was living on the fringes.
References:
Markow, T., & O'Grady, P. (2008). [Reproductive ecology of _Drosophila_](http://biology.ucsd.edu/labs/markow/articles/Markow%20and%20OGrady%202008.pdf). _Functional Ecology_ **22**, 747-759.
Simpson, P. (2002). [Evolution of development in closely related species of flies and worms](http://www.nature.com/nrg/journal/v3/n12/abs/nrg947.html). _Nat Rev Genet_ **3**:907-917.
Stark, A., _et al_. (2007). [Discovery of functional elements in 12 _Drosophila_ genomes using evolutionary signatures](http://www.nature.com/nature/journal/v450/n7167/full/nature06340.html). _Nature_**450**, 219-232.
