Reconstruction of Ancestral Proteins
Yesterday a really cool paper came out in the journal Nature that demonstrates why evolutionary theory is so useful and fruitful in biology. A team of researchers has recreated an ancestral bacterial protein to determine that the ancestral bacteria grew in hot water around 3.5 billion years ago.
 How can you reconstruct the protein sequence of organisms long gone? Using evolutionary theory, you can take the genomes of existing related organisms, and work backwards to the sequence of their common ancestor. This has been done to reconstruct ancestral steroid receptors, archosaur visual pigments and ancient coral fluorescent proteins. Of course, there are several technical issues involved in reconstructing the phylogeny and the ancestral sequence. You have to have the right phylogeny to construct the gene of the ancestral organism, and there are a number of issues that revolve around the rate of nucleotide replacement and biases in the frequencies of amino acids over time. However, these can be addressed in a number of ways, including using powerful statistical techniques and testing alternative phylogenies.
How can you reconstruct the protein sequence of organisms long gone? Using evolutionary theory, you can take the genomes of existing related organisms, and work backwards to the sequence of their common ancestor. This has been done to reconstruct ancestral steroid receptors, archosaur visual pigments and ancient coral fluorescent proteins. Of course, there are several technical issues involved in reconstructing the phylogeny and the ancestral sequence. You have to have the right phylogeny to construct the gene of the ancestral organism, and there are a number of issues that revolve around the rate of nucleotide replacement and biases in the frequencies of amino acids over time. However, these can be addressed in a number of ways, including using powerful statistical techniques and testing alternative phylogenies.
Gaucher, Govindarajan and Ganesh examined the thermostability of Elongation factor Tu (EF Tu) proteins reconstructed by phylogenetic analysis. EF Tu is critical in protein synthesis, and there are lots of EF Tu sequences available to build reasonably reliable phylogenies. Importantly, the stability of Ef Tu is known to correlate with environmental temperature in extant organisms. So the stability of reconstructed EF Tu should give us a readout of the environmental temperature at the time the ancestral organism lived.
The authors were able to reconstruct the EF Tu proteins at several nodes in the phylogenetic tree, corresponding to different times between 500 million years ago and 3.5 billion years ago. The reconstructed proteins show a progressive increase in thermostability, suggesting that bacteria living around 3.5 billion years ago were living in very hot water. This analysis can’t determine whether the organisms were living it hot springs, or if the entire oceans were hot, but the thermostability of the proteins correlate strikingly with oceanic temperatures inferred from oxygen and silicon isotope ratios.
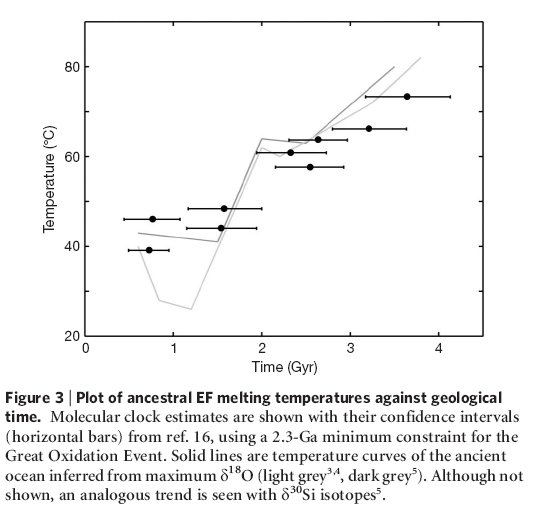 Figure from Gaucher et al, Nature (2008) 451, 708
Figure from Gaucher et al, Nature (2008) 451, 708
This analysis, deeply rooted in evolutionary concepts, has allowed us to infer something important about the environments of ancient creatures. Also, by generating functional proteins, we can better understand the mechanisms of the enzymes themselves. Contrast this with Intelligent Design, which has yet to make a useful prediction of any kind.
Also note that functional proteins were produced. This is also true of the reconstructed ancestral steroid receptors, archosaur visual pigments and ancient coral fluorescent proteins. The Intelligent Design movement in general has wedded itself to the idea that functional proteins are “extremely isolated” and that you require an intelligent designer to get from one isolated island of function to another. Thus, according to ID, you cannot infer the ancestral structure of a protein from existing organisms. The successful reconstruction of multiple, function proteins by evolutionary phylogenetics represents a major blow to this ID concept.
- Eric A. Gaucher, Sridhar Govindarajan & Omjoy K. Ganesh (2008) Palaeotemperature trend for Precambrian life inferred from resurrected proteins Nature 451: 704-707
- Thornton JW, Need E, Crews D (2003) Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 301: 1714–1717.
- Chang BS, Jonsson K, Kazmi MA, Donoghue MJ, Sakmar TP (2002) Recreating a functional ancestral archosaur visual pigment. Mol Biol Evol 19: 1483–1489.
- Chang BS, Ugalde JA, Matz MV (2005) Applications of ancestral protein reconstruction in understanding protein function: GFP-like proteins. Methods Enzymol 395: 652–670.
