ERV & HIV versus Behe. Behe loses.
by Abbie Smith
Editor’s Note: This is a guest post by Abbie Smith, the blogger behind the Endogenous Retrovirus (ERV) blog (original post), who we marked early as having a special talent for this creationism-rebutting stuff. Abbie works in an HIV lab and has a few things to say about Behe’s argument in_The Edge of Evolution_that evolution hasn’t/can’t produced any novel adaptations, genes, or protein-protein binding sites during the evolution of HIV, the virus that causes AIDS, unless you are one of the several Discovery Institute fellows who denies this along with denying evolution.
Oh, and speaking of the Discovery Institute and Behe, Behe is apparently going to be on the Colbert Report tonight. Colbert has been setting an excellent example by making a point of it to bring scientists on his show – Kenneth Miller, Neil DeGrasse Tyson, Shubin I think. Hopefully Colbert knows that Behe isn’t quite like these other guys. I recall that Dembski claimed he was sick after he appeared on The Daily Show next to some weird new-ager in 2005.
Anyway, if Colbert doesn’t hit the appropriate snark mark tonight, you’ll get it from Abbie for sure. I thought I got Behe pretty good in Of Cilia and Silliness, but this _really takes the cake._
—Nick Matzke
Michael Behe, please allow me to introduce myself…
I’m ERV. This is my dog, Arnold Schwarzenegger. And this is my friend, Vpu. I presume you and Vpu haven’t met, as you recently repeated in an interview with World magazine the same sentiment you gurgled ad nauseam in ‘Edge of Evolution’:
“Like malaria, HIV is a microbe that occurs in astronomical numbers. What’s more, its mutation rate is 10,000 times greater than that of most other organisms. So in just the past few decades HIV has actually undergone more of certain kinds of mutations than all cells have endured since the beginning of the world. Yet all those mutations, while medically important, have changed the functioning virus very little. It still has the same number of genes that work in the same way. There is no new molecular machinery. If we see that Darwin’s mechanism can only do so little even when given its best opportunities, we can decisively conclude that random mutation did not build the machinery of life.”
I find it rather difficult to believe that you two haven’t crossed paths, as Vpu turns up in a simple Google search. And as a matter of fact, Vpu is sitting right there in the totally unnecessary and worthless diagram in ‘Edge of Evolution’. See? Right there:

If you had taken the time to label this pointless diagram, you might have noticed your error, and we wouldn’t need to have this conversation. Alas, Vpu is a tiny molecule, and he’s easy to overlook if you don’t want to see him.
Vpu is, in fact, a new gene.1 Of the five major phylogenetic groups of SIV, Vpu is only found in one group– Chimpanzee SIV (SIVcpz) and its descendants – including HIV-1. It is absent in all of the other major lineages (Sooty Mangabey, African Green Monkey, Sykes Monkey, and L’Hoest Monkey). This means that Vpu is in HIV-1 but not HIV-2.2
Ah, Michael Behe, you might try to talk your way around Vpu now (though you were evidently unaware of its existence moments ago) by insisting that it is not *new* new. “Sure it’s new in chimpanzees, but its not *new* in HIV-1!” Sorry, you’ll find no escape with that limp-wristed, ad hoc parry. SIVcpz Vpu and HIV-1 Vpu act in different ways, biochemically, which is predictable enough when you do something as simple as comparing amino acid sequences. For instance, if you compare a laboratory strain gag to SIVcpz gag, you get a similarity of ~75%.3 Not too shabby. On the other hand, if you compare the subunit portion of env (the gene I use to create phylogenetic trees because it’s the most variable between viruses) you get an AA similarity of only ~59.5%.
The amino acid similarity between HIV-1 Subtype B Vpu and SIVcpz Vpu is ~37%. Ah but that study was published in 1990. Perhaps things are different now? I found the AA sequence of NL4-3 (lab standard Subtype B) and several recently entered SIV cpz sequences at the Los Alamos National Laboratory HIV Sequence Database4 – I got the same numbers. Highest was ~39% AA sequence similarity.
Turns out a LOT of evolution has been going on in HIV-1 since it was transferred to humans 50-60 years ago. What are the biochemical implications of these differences?
In humans, there are two functions of Vpu5 – one is inducing the degradation of CD4 molecules. CD4 is the host cell receptor HIV needs for infection. Removing CD4 from the cell surface prevents superinfection (more than one virus infecting the same cell) and helps prevent newly released viruses from turning around and infecting the same cell (also prevented by an HIV maturation step involving protease). To put this the simplest way possible, Vpu involves the evolution of at least two protein-protein interaction sites – one to interact with CD4, one to interact with the pathway that degrades the CD4.
The second function is to act as an ion channel in the host cell plasma membrane.6 Five Vpu proteins come together to form a Na+K+ viroporin.7 This has been shown to assist in particle release, making the cell charge more conducive to the release of new particles. This involves the evolution of more protein-protein interactions – the individual Vpu proteins must interact to form the pentamer, plus an action site that can be used to block ion flow.8
Viroporin capabilities have not been found with SIVcpz Vpu. Knowing what we know about Vpu, this is not surprising. If you scramble the transmembrane region of HIV-1 Vpu (the portion responsible for the ion channel formation), viral release is crippled.9 And when you compare AA homology between SIVcpz and HIV-1 Vpu in the transmembrane region is unremarkable (roughly two) – that’s as good as a ‘scramble’. So theoretically, ion channel formation evolved in HIV-1 when it infected humans to overcome a species specific and cell specific host factor.10 Though the list of viroporins discovered is continually growing, the evolution of a viroporin de novo is not menial task.
This seems like a pretty significant biochemical change in HIV-1, to me.
But the ‘pathetic’ evolution doesn’t stop there. The feature both Vpus have in common, CD4 degradation, is carried out in completely different ways. HIV-1 Vpu requires two casein kinase II sites. You could almost call it irreducibly complex – if you dont have both CKII sites, CD4 isn’t degraded. Yet some SIVcpz Vpus have only one CKII site, and instead utilize a simple string of negatively charged amino acids in place of the second site.11 Different ways of performing similar tricks with totally different amino acids. I think that’s biochemically significant as well.
Ah, Michael Behe, you might try to talk your way around Vpu NOW by saying, “Vpu might be *new* new in HIV-1, but its not *NEW* *new* new. It hasnt changed in HIV-1 since humans acquired it!”
Alas, ‘same number of genes that work in the same way’ goes beyond the differences between HIV-1 Vpu and SIVcpz Vpu. HIV-1 is divided into three groups, M, N, O. Group M is the one making a mess of the world right now, and is further divided into Subtype A, B, C, etc, and circulating recombinant forms of the subtypes (Subtype AG, for instance). Two relatively well characterized subtypes are Subtype B and C. Subtype C HIV can be defined by its Vpu, as it is so different from the other subtypes.12
For instance, Subtype C Vpus are characteristically longer than the others, have key phosphorylation sites shifted, have an extra CKII site, and its tertiary structure is totally different (Subtype B Vpus have an Mr of 43,000 in an SDS-PAGE gel, while Subtype C is 34,000). But what does this mean, biochemically?
It turns out that one of the biochemical differences is that Subtype B Vps have a Golgi retention signal in the second alpha-helix of the cytoplasmic domain.13 This means that Subtypes B Vpu prefers (if you will excuse me personifying a virus) to be in the Golgi, helping degrade CD4, while Subtype C Vpu prefers to be in the plasma membrane, assisting with the release of new viruses. Michael Behe, if you don’t understand the epidemiological and clinical significance of this ‘pathetic’ evolution, well, that might explain why you aren’t doing HIV research.
So here’s a quick time-line for the evolution of impossible genes and impossible protein-protein interactions, courtesy of Ian Musgrave (one of those dreadful PhDs/Professors that has something to teach this pre-grad student):
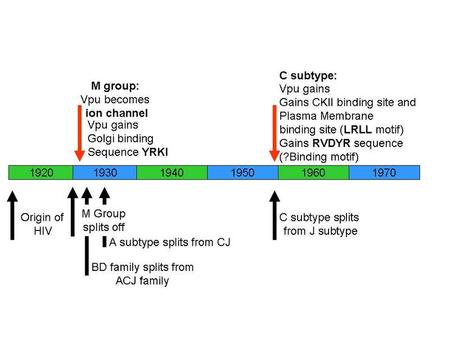
Michael Behe, as a courtesy to you, this essay isn’t even going to touch Vpx, a gene specific to the HIV-2 and SIV sm lineage. Vpx is one of those pesky gene duplications you say don’t exist in HIV (Vpr x2). I also won’t point out how silly it is of you to claim HIV has not evolved biochemically, when the HIV research community has barely scratched the surface of HIV’s biochemical evolution (for the love of Pete, we don’t even know what HIV-1 Subtype Cs gp120-gp41 look like!).
Look, the fact of the matter is, all of this information on Vpu is publicly available. No one was hiding this information. This wasn’t a ‘trick.’ Vpu was not discovered yesterday – it was discovered in 1988. There is no excuse for you to write an entire book on the premise of HIV not being able to do something, when it is clear that these impossible feats did happen. This is just one of a billion plus examples of lazy Creationists taking advantage of the ignorance of their followers. I’m just a friggen pre-grad student who knew what the HIV-1 genome looked like and had a few minutes to do a PubMed search. I haven’t even taken a course in biochemistry.
*sigh* So let’s hear it, Behe, what’s your excuse for missing this? Go ahead – run off to PubMed and find a paper to pubjack. I’m not going anywhere.
References:
- A novel gene of HIV-1, vpu, and its 16-kilodalton product
- Diversity and Evolution of Primate Lentiviruses
- Genetic organization of a chimpanzee lentivirus related to HIV-1
- LANL HIV Sequence Database
- The HIV-1 Vpu protein: a multifunctional enhancer of viral particle release
- The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release
- The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels
- Drug-protein interaction with Vpu from HIV-1: proposing binding sites for amiloride and one of its derivatives
- Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells
- HIV-1 Vpu Promotes Release and Prevents Endocytosis of Nascent Retrovirus Particles from the Plasma Membrane
- Vpu-mediated CD4 down-regulation and degradation is conserved among highly divergent SIV(cpz) strains
- Molecular characterization of the HIV type 1 subtype C accessory genes vif, vpr, and vpu
- Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that is responsible for retention in the golgi complex and its absence in the Vpu protein from a subtype C HIV-1
